Methods
pycisTarget is a motif enrichment suite that combines different motif enrichment approaches such as cisTarget and Homer; and a novel approach to compute Differentially Enriched Motifs between sets of regions called DEM. Pycistarget is available at https://github.com/aertslab/pycistarget.
Homer
PycisTarget includes a wrapper to run Homer’s findMotifsGenome.pl, allowing the identification of known and de novo motifs (by default using default Homer parameters). For identifying cistromes for each motif, found motifs are used as input for homer2 find. Known motifs are annotated according to the motifs annotation in the SCENIC+ motif collection. To annotate de novo motifs, tomtom is run with the SCENIC+ motif collection to identify the closest match, allowing to transfer its annotation to the de novo motif when specified. To form TF cistromes, motif-based cistromes are combined based on the TF annotations.
Generation of cisTarget databases
cisTarget and DEM require ranking and score-based databases, respectively, with regions as rows, motifs (or motif clusters) as columns, and motif enrichment scores, or ranking values of these scores, as values. For this, we have developed an adapted Cluster-Buster115, which is now 2 times faster. Cluster-Buster uses Hidden Markov Models (HMMs) to score clusters of motifs (i.e., Cis-Regulatory Modules (CRM)) given a set of regions. This implementation is available at https://resources.aertslab.org/cistarget/programs/cbust. Briefly, given a motif collection (in cb format) and a set of regions, we run Cluster-Buster using each motif across all regions. When dealing with motif clusters, Cluster-Buster uses all motif variations by implanting each motif as a hidden state in a HMM, and each candidate sequence receives a log-likelihood ratio (LLR) score per motif cluster (i.e. CRM score). A scores database is first generated by taking the highest CRM score per sequence. A ranking database is then generated by ranking, for each motif, all the regions by decreasing motif score. The code and documentation to generate these databases is available at https://github.com/aertslab/create_cisTarget_databases. In addition, we provide precomputed databases using predefined sets of regulatory regions for hg38, mm10 (using SCREEN regions) and dm6 (using cisTarget regions, defined by partitioning the entire non-coding Drosophila genome based on cross-species conservation) at: https://resources.aertslab.org/cistarget/ .
cisTarget
pycisTarget implements our ranking based motif enrichment method cisTarget. Briefly, we first score genomic regions (i.e. consensus peaks, or predefined regions from SCREEN) using a motif collection with Cluster-Buster, as previously described. These regions will be ranked based on their motif score for each motif. The input regions are intersected with regions in the database (with at least 40% overlap). cisTarget uses a recovery curve approach (for each motif), in which a step is taken in the y-axis when as region in the motif ranking (x-axis) is found in the region set. The Area Under the Curve for each motif is normalized based on the average AUC for all motifs and their standard deviation, resulting in a Normalized Enrichment Score (NES) that is used to quantify the enrichment of a motif in a set of regions. By default, motif that obtain a NES above 3.0 are kept. To obtain the target regions for each TF, the motif-based cistromes of motifs annotated to that TF are combined.
DEM
DEM performs a Wilcoxon test between scores in foreground and background region sets to assess motif enrichment. Briefly, we first score genomic regions (i.e. consensus peaks, or predefined regions from SCREEN) using a motif collection with Cluster-Buster, as previously described. Regions in the input region sets are intersected with regions in the database (with at least 40% overlap), and the Wilcoxon test is performed between CRM score distributions in the two groups. By default, motifs with adjusted p-val < 0.05 (Bonferroni) and Log2FC > 0.5 are kept. A cistrome for each motif is generated by simultaneously optimizing precision and recall to separate foreground from background regions or using a predefined CRM threshold. To obtain the target regions for each TF, the motif-based cistromes of motifs annotated to that TF are combined.
SCENIC+
Within the SCENIC+ workflow, motif enrichment is performed by default in binarized topics and DARs calculated by pycisTopic, using cisTarget and DEM (including and excluding promoters from the region sets). By default, DEM is run using topics or DARs as foreground and 500 regions in other topics/DARs as background (with the same proportion of promoters as in the foreground). Additional region sets (e.g. DARs derived from specific contrasts instead of using all populations as background) can be easily added. Cistromes derived from all the motif enrichment analysis are merged by TF to generate a final set of TF-region cistromes.
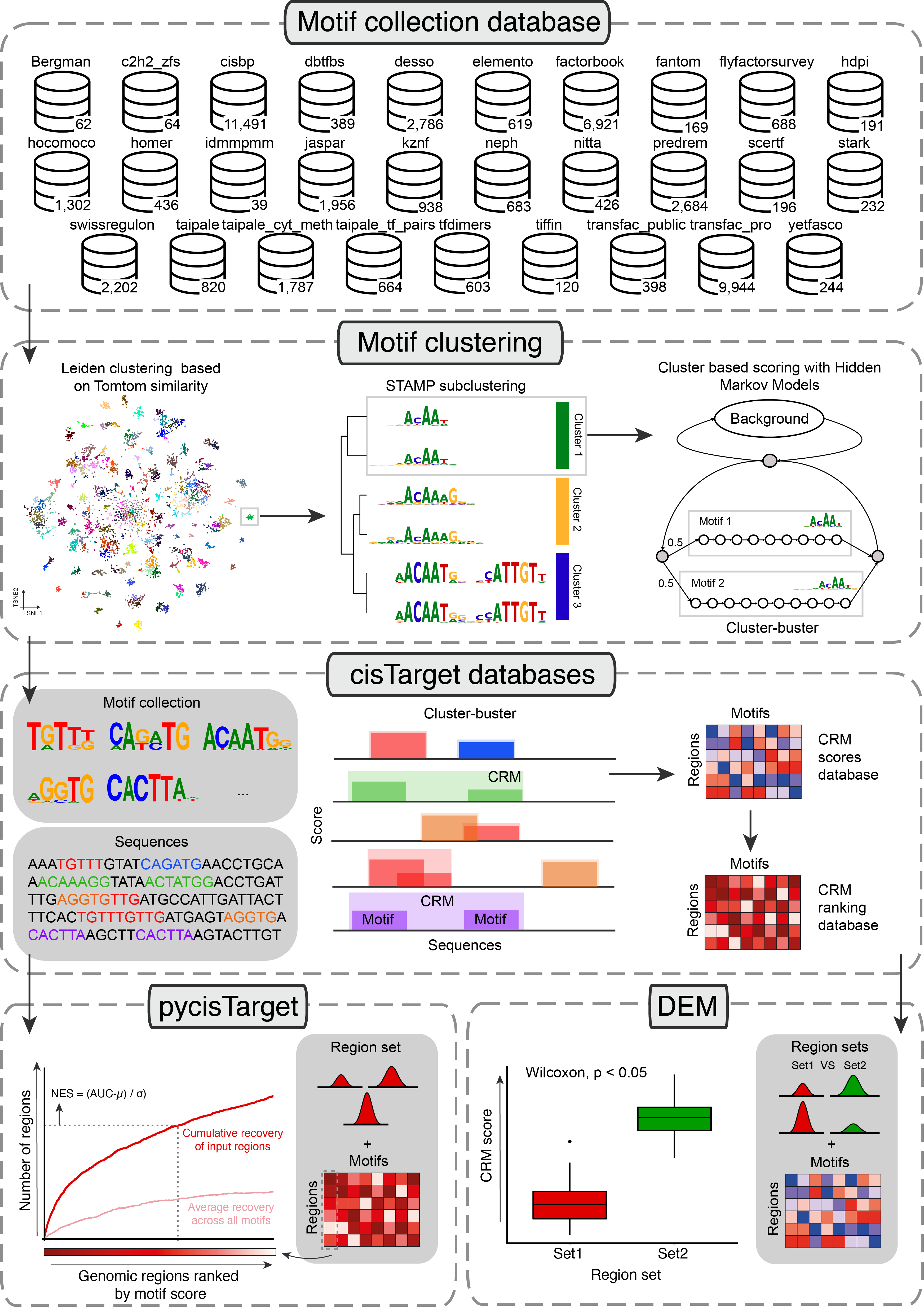
The SCENIC+ motif collection includes 34,524 unique motifs gathered from 29 motif collections, which were clustered with a two-step strategy in which motifs are first clustered using Leiden clustering on the -log10 Tomtom q-value matrix, and these clusters are then refined using STAMP. Region scoring is performed using Hidden Markov Models (HMMs) models with an updated version of Cluster-Buster, in which clusters are scored using all the motifs as states instead of using a consensus motif. Ranked scores are used as input for pycisTarget, in which a recovery curve is built for each motif and region set. The Normalized Enrichment Score (NES) indicates how enriched a motif is in the region set. Scores are used for Differentially Enriched Motifs (DEM) analysis, in which a Wilcox test is performed between foreground and background region sets to assess enrichment.